Phage Genome Annotation
Once you have identified the high-quality and complete genomes from the CheckV results, you can annotate them using a tool such as pharokka. The following sections will walk you through how to setup and run pharokka.
Installing pharokka
The recommended way to install pharokka is using conda.
# Create a new conda environment and install pharokka
conda create -n pharokka -c bioconda pharokka
# Activate pharokka conda environment
conda activate pharokka
Download and install the pharokka databases
install_databases.py -o <path/to/databse_dir>
Running pharokka
Here is an example command to run pharokka on the complete and high-quality resolved genomes.
pharokka.py -i complete_hq_genomes.fasta -o pharokka_output -t 16 -d <path/to/database_dir>
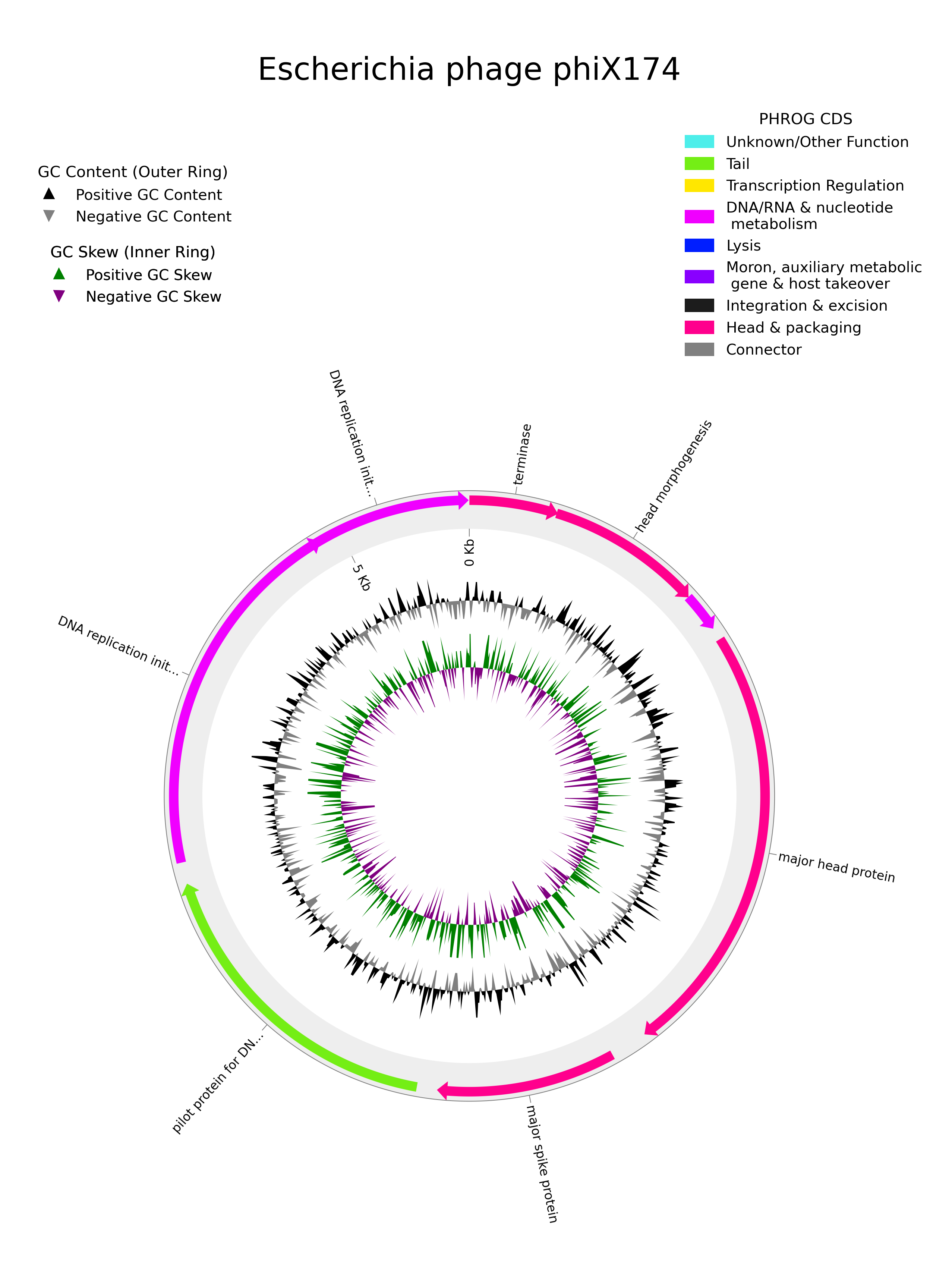
Circular genome plot
You can use the pharokka_plotter.py implementation from pharokka to create circular genome plots with annotations.
Let's assume that you have already run pharokka on all of the complete and high-quality resolved genomes and the output is available in pharokka_output. You can pick one genome to plot. For example, let's consider the genome phage_comp_280_cycle_1.fasta which is phiX174.
We start by reorienting the genome to start from the terminase large subunit. You can look up the starting position and strand of the terminase large subunit from the output file pharokka_output/pharokka_cds_final_merged_output.tsv. For example, let's take the starting position as 617 on the positive strand. You can run pharokka again for this genome with reorientation as follows.
pharokka.py -i resolved_phages/phage_comp_280_cycle_1.fasta -o pharokka_output_phage_comp_280_cycle_1 -d <path/to/databse_dir> -t 16 --terminase --terminase_strand 'pos' --terminase_start 617
Then you can run the plotting command as follows.
pharokka_plotter.py -i resolved_phages/phage_comp_280_cycle_1.fasta -n phage_comp_280_cycle_1_plot -o pharokka_output_phage_comp_250_cycle_1 -t "Escherichia phage phiX174"